Mayer Lab – Research
At the core of our research lies the question of how cells ensure that the required RNA is produced in the correct amounts at the right times to control cellular functions. Specifically, our research aims to understand the molecular mechanisms that control the synthesis of nascent RNA by RNA polymerase II (Pol II) and RNA processing — the two key steps that lead to functional RNAs. We primarily study these fascinating molecular processes within the native chromatin environment of mature and differentiating mammalian cells, including patient cells. Our research aims to improve our basic understanding of these fundamental processes, as well as investigating how nascent transcription and RNA processing underlie cell differentiation and how dysfunction can lead to human pathology. To address these questions, we have developed and are applying a high-resolution, functional, multi-omics approach incorporating transcriptome-wide experimental methods, cell line engineering, super-resolution microscopy, and computational tools.
Molecular mechanisms of gene expression regulation
The genomes of mammals are extensively transcribed by RNA polymerase II (Pol II), resulting in a wide variety of protein-coding and non-coding RNAs. Our research focuses on revealing the molecular mechanisms that control genome-wide RNA synthesis by Pol II and RNA processing — the two main steps that yield functional RNAs. We also investigate the fascinating crosstalk between transcription and RNA processing within the native chromatin environment of mammalian cells. Our research aims to identify the factors that regulate and coordinate these two fundamental processes, which are at the basis of cell function.
Our research considers that genes are controlled not only by promoters, but also by distal regulatory elements known as enhancers. Furthermore, transcription is not restricted to genes; it is also common in the non-coding part of the genome, particularly at enhancers, where it gives rise to enhancer RNAs (eRNAs). We have recently obtained an almost complete map of transcribed enhancers and nascent eRNAs in human leukemia cells (Bressin et al., Nature Communications, 2023). Our research focuses on how enhancers communicate with their target genes to regulate RNA levels (Altendorfer et al., Nature Structural & Molecular Biology, 2025). We also investigate this subcellular communication in the context of biomolecular condensates in cells.
To gain new insights into the fundamental processes underlying the regulation of gene expression and its dysregulation in human malignancies, we employ an interdisciplinary approach combining functional genomics, systems biology, biochemistry, microscopy and advanced computational methods. We also develop new experimental methods, such as protocols for high-resolution genome-wide nascent RNA analysis, and computational tools.
Cell differentiation – understanding regulatory dynamics
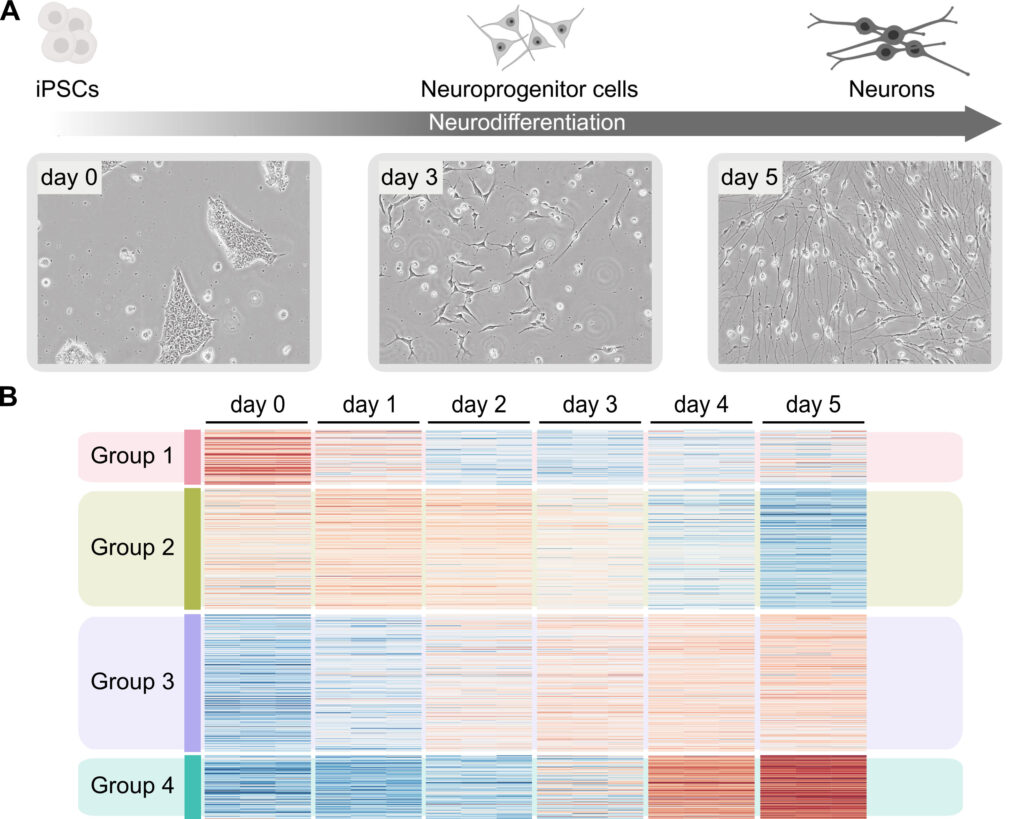
The human body consists of over 200 different cell types that perform various functions. These cell types emerge through cell differentiation — the fundamental process by which unspecialized cells, such as stem cells, develop into specialized cells with distinct structures and functions, underpinned by distinct gene expression profiles. But how are cell-type-specific gene expression programs established and maintained? What molecular mechanisms control the dynamic changes in RNA and proteins as cells transition from a pluripotent to a differentiated state? How are changes in cell state during differentiation regulated? To address these questions, we are adapting and improving cellular models for the study of human cell differentiation ex vivo. Using these models, we aim to identify the determinants and molecular mechanisms of cell state transitions, including chromatin and transcriptional dynamics, during differentiation. Our focus at the moment is on human neuronal differentiation (neurogenesis).
From understanding molecular mechanisms to understanding disease
We use our integrative, interdisciplinary approach, drawing on our knowledge of molecular mechanisms, to elucidate how changes in gene expression — particularly transcription and RNA processing — cause disease. Our ultimate goal is that the new insights into disease mechanisms will help to improve diagnosis and open up new avenues for more specific therapeutic interventions in the future. We aim to understand the pathological mechanisms by creating cellular disease models using cutting-edge cell engineering techniques. One of our current focuses is Coffin-Siris syndrome, a neurological disorder caused by mutations in the BAF chromatin remodelling complex, which interacts with the RNA polymerase II transcription machinery. Our other focus is on the molecular mechanisms of leukemia. In this area, we are seeking to clarify the specific role of BET family proteins in different leukemias, such as acute myeloid leukemia (AML) and chronic myelogenous leukemia (CML).